






 |
|---|
| Mint production near St. Johns, Michigan provides spearmint (shown here) and peppermint essential oils to flavor many foods and other commercial products. © 2015 Grant Godden. |
Evolution of specialized metabolite biosynthetic pathways in the Lamiaceae: Sources of chemical diversity for molecules essential for human use and plant defense
Plant species exhibit robust diversity of secondary metabolites. We hypothesize that plant chemical diversity can be enhanced or modified as a result of gene and/or genome duplication along with subsequent evolution that leads to divergence of genes involved in the biosynthesis, regulation, and compartmentalization of secondary metabolites. The Lamiaceae, or mint family, is an excellent system to test this hypothesis, as plant species in this family display high chemical diversity that is a central feature of their agricultural value as food and industrial additives, herbs, ornamentals, and medicines. Integration of genome, transcriptome, and metabolome datasets in phylogenetic and evolutionary analyses will permit elucidation of not only key evolutionary events but also the genes that contribute to the phenotypic diversity observed in Lamiaceae species.
I. OBJECTIVES
Our overall objective is to determine how evolution led to a collection of genes and regulatory features responsible for the immense and economically important chemodiversity in the Lamiaceae, which may be leveraged for new economic uses. We have three inter-related specific scientific objectives, along with two educational objectives:
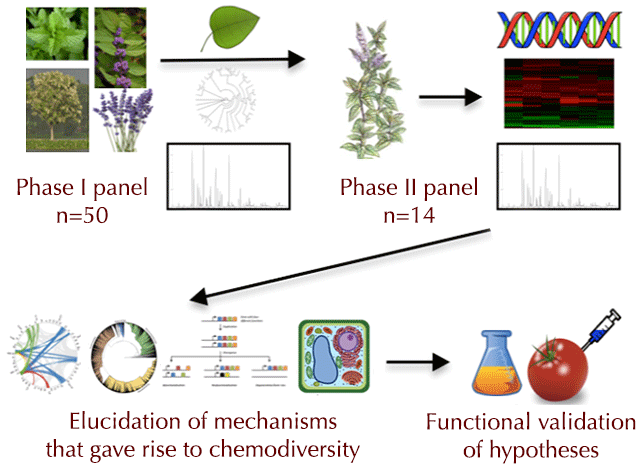
Obj. 1. Perform transcript and metabolite profiling in a panel of 50 phylogenetically diverse species of Lamiaceae to identify a subset of 14 species that provides a robust representation of genetic and chemical diversity in this family (see Figure 1: Phase I below).
Obj. 2. Determine the genetic and evolutionary basis of chemodiversity by integrating genome, expression, metabolite, and phylogenetic datasets from 14 diverse Lamiaceae species (see Figure 1: Phase II below).
Obj. 3. Decipher the biochemical and molecular basis for metabolic diversification of monoterpenes and iridoids in the Lamiaceae (see Figure 1: Phase II below).
Obj. 4. Provide interdisciplinary training of high school, undergraduate, graduate students and postdoctoral fellows that integrates biochemistry, bioinformatics, evolution, genomics, and phylogenetics. Broaden participation of under-represented groups throughout the project.
Obj. 5. Engage the public, including K-5 children, in phenotypic diversity, including chemical diversity, in plants through activities at the Michigan State University (MSU) 4-H Children’s Garden and the Florida Museum of Natural History at the University of Florida (UF). Read more >
 |
|---|
| Figure 1. Workflow of the Mint Genome Project. A screen of leaf monoterpene and iridoid profiles in 50 Phase I species will be coupled with transcriptome sequencing and phylogenetic analyses to identify a robust set of 14 species (Phase II) for detailed analyses. Work with the Phase II species will include genome sequencing, expression and metabolite profiling in a broad set of tissues, and phylogenetic, evolutionary, and comparative genomic analyses. This will lead to elucidation of evolutionary mechanisms such as whole genome and gene duplications, divergence and selection, neo- and sub- functionalization, and differential subcellular localization that gave rise to the extant chemodiversity observed in Lamiaceae. Functional characterization of a subset of biosynthetic enzymes in heterologous expression systems will provide evidence for these mechanisms. |
Research Team
This project brings together scientists with complementary research expertise, with documented strengths in plant genomics/bioinformatics (Buell), phylogenetics/evolution (P Soltis, D Soltis), and secondary metabolites (Dudareva and O’Connor). Buell has been involved in genome and transcriptome sequencing projects since 1999 and has extensive experience in annotating genomes and disseminating data to the research community. Both P Soltis and D Soltis have over 25 years expertise in plant phylogenetics as well as recent use of genomic datasets for evolutionary analyses. Dudareva has 20 years experience in plant biochemistry, specializing in the biosynthesis and regulation of primary and secondary metabolites including terpenoids. O’Connor has 15 years experience in organic chemistry and has been seminal in discovery of iridoid biosynthetic pathway components in plants. For more information about the full research team, follow the link below.
| Overview |
| Project Description |
| Taxonomic Sampling |
© 2015 Univeristy of Georiga
Buell Lab | Department of Crop and Soil Sciences, 111 Riverbend Rd., Athens, GA 30602 | Tel. (706) 542-6227